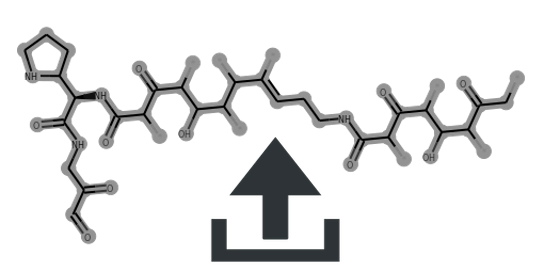
Cluster scaffolds:
2 |
1 |
Putative metabolites:
| Name | Rank | Metabolite Score | Similarity | MCS Score | Post-Mod Score | Post-Mod Info | Source | Link | Smiles | Molview | Full Name | Figure | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Taxlllaid | 0 | 0.62 | 0.59 | 0.8 | 0.0 | 5-Ring:0/1 | MIBiG | Source | CC(C)CCCC(=O)N[C@@H]1C(=O)NC(C)C(=O)N[C@@H](CC(C)C)C(=O)NC(CC(C)C)C(=O)N[C@H](CC(C)C)C(=O)N[C@@H](CC(C)C)C(=O)N[C@@H](C)C(=O)O[C@@H]1C | view | Taxlllaid | ||
| WLIP | 1 | 0.58 | 0.57 | 0.73 | 0.0 | 5-Ring:0/1 | MIBiG | Source | CCCCCCC[C@@H](O)CC(=O)N[C@@H](CC(C)C)C(=O)N[C@H](CCC(=O)O)C(=O)N[C@H]1C(=O)N[C@H](C(C)C)C(=O)N[C@H](CC(C)C)C(=O)N[C@H](CO)C(=O)N[C@@H](CC(C)C)C(=O)N[C@H](CO)C(=O)N[C@@H]([C@@H](C)CC)C(=O)O[C@@H]1C | view | WLIP | ||
| cyclomarin D | 2 | 0.57 | 0.57 | 0.72 | 0.0 | 5-Ring:0/1 | MIBiG | Source | C=CC(C)(C)n1cc([C@@H](O)[C@@H]2NC(=O)[C@H]([C@H](C)C=C(C)C)NC(=O)[C@H](CC(C)C)N(C)C(=O)[C@H](C(C)C)NC(=O)[C@H]([C@H](OC)c3ccccc3)NC(=O)[C@H](C)NC(=O)[C@H](C[C@@H](C)CO)NC2=O)c2ccccc21 | view | cyclomarin D | ||
| Xenotetrapeptide | 3 | 0.57 | 0.59 | 0.69 | 0.0 | 5-Ring:0/1 | MIBiG | Source | CC(C)C[C@@H]1NC(=O)[C@@H](C(C)C)NC(=O)[C@H](C(C)C)NC(=O)[C@@H](C(C)C)NC1=O | view | Xenotetrapeptide | ||
| Myxochromide S1 | 4 | 0.56 | 0.57 | 0.69 | 0.0 | 5-Ring:0/1 | MIBiG | Source | C/C=C/C=C/C=C/C=C/C=C/C=C/C=C/C(=O)N(C)C1C(=O)NC(CC(C)C)C(=O)NC(C)C(=O)NC(C)C(=O)NC(CCC(N)=O)C(=O)CC1C | view | Myxochromide S1 |