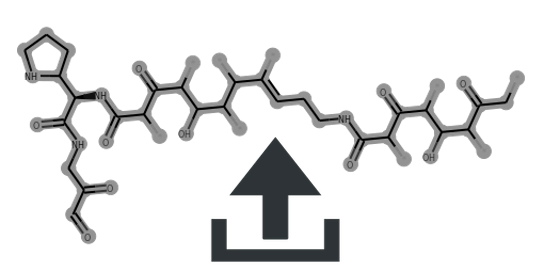
Cluster scaffolds:
1 |
Putative metabolites:
| Name | Rank | Metabolite Score | Similarity | MCS Score | Post-Mod Score | Post-Mod Info | Source | Link | Smiles | Molview | Full Name | Figure | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E-837 | 0 | 0.53 | 0.55 | 0.65 | 0.0 | Glyco:0/3 | MIBiG | Source | C/C=C(\C)C(O)C(C)/C=C/C=C(\C)CCC(O)CC1=C(C)C(=O)C(C)(O)O1 | view | E-837 | ||
| E-492 | 1 | 0.51 | 0.55 | 0.6 | 0.0 | Glyco:0/3 | MIBiG | Source | C/C=C(\C)C(O)C(C)/C=C/C=C/CCC(O)CC1=C(C)C(=O)C(O)(C(C)O)O1 | view | E-492 | ||
| Aurafuron A | 2 | 0.5 | 0.55 | 0.58 | 0.0 | Glyco:0/3 | MIBiG | Source | CC1=C(CC(O)/C=C\C=C\C(C)C(O)/C(C)=C/CC(C)C)OC(C)(O)C1=O | view | Aurafuron A | ||
| E-975 | 3 | 0.49 | 0.52 | 0.59 | 0.0 | Glyco:0/3 | MIBiG | Source | CC/C=C(\C)C(O)C(C)/C=C/C=C/CCC(O)CC1=C(C)C(=O)C(O)(C(C)O)O1 | view | E-975 | ||
| AF-toxin | 4 | 0.44 | 0.54 | 0.47 | 0.0 | Glyco:0/3 | MIBiG | Source | CCC(C)C(OC(=O)C(O)C(C)(C)O)C(=O)OC(/C=C/C=C/C=C/C(=O)O)C1(C)CO1 | view | AF-toxin |