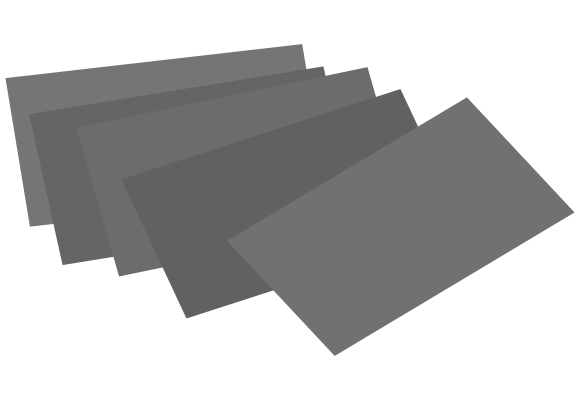
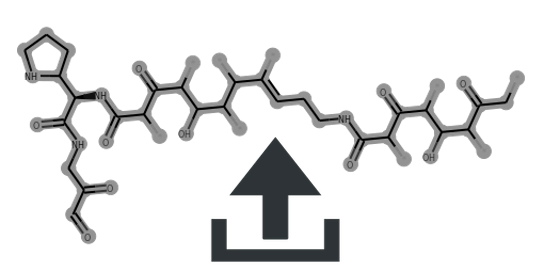
Cluster scaffolds:
1 |
Putative metabolites:
| Name | Rank | Metabolite Score | Similarity | MCS Score | Post-Mod Score | Post-Mod Info | Source | Link | Smiles | Molview | Full Name | Figure | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xenortide A | 0 | 0.56 | 0.59 | 0.67 | 0.0 | Glyco:0/3 | MIBiG | Source | CN[C@@H](CC(C)C)C(=O)N(C)[C@@H](Cc1ccccc1)C(=O)NCCc1ccccc1 | view | Xenortide A | ||
| Xenortide C | 1 | 0.55 | 0.59 | 0.65 | 0.0 | Glyco:0/3 | MIBiG | Source | CN[C@H](C(=O)N(C)[C@@H](Cc1ccccc1)C(=O)NCCc1ccccc1)C(C)C | view | Xenortide C | ||
| heterobactin A | 2 | 0.51 | 0.53 | 0.62 | 0.0 | Glyco:0/3 | MIBiG | Source | N/C(=N\C(=O)c1cccc(O)c1O)NCCC[C@@H](NC(=O)c1cccc(O)c1O)C(=O)NCC(=O)N[C@H]1CCCN(O)C1=O | view | heterobactin A | ||
| Xenortide B | 3 | 0.5 | 0.53 | 0.61 | 0.0 | Glyco:0/3 | MIBiG | Source | CN[C@@H](CC(C)C)C(=O)N(C)[C@@H](Cc1ccccc1)C(=O)NCCc1c[nH]c2ccccc12 | view | Xenortide B | ||
| Xenortide D | 4 | 0.5 | 0.53 | 0.6 | 0.0 | Glyco:0/3 | MIBiG | Source | CN[C@H](C(=O)N(C)[C@@H](Cc1ccccc1)C(=O)NCCc1c[nH]c2ccccc12)C(C)C | view | Xenortide D |