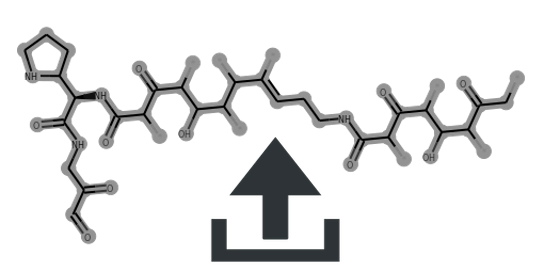
Cluster scaffolds:
1 |
Putative metabolites:
| Name | Rank | Metabolite Score | Similarity | MCS Score | Post-Mod Score | Post-Mod Info | Source | Link | Smiles | Molview | Full Name | Figure | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tabtoxin | 4 | 0.61 | 0.46 | 0.62 | 1.0 | Non detected. | MIBiG | Source | C[C@@H](O)[C@H](NC(=O)[C@@H](N)CC[C@]1(O)CNC1=O)C(=O)O | view | tabtoxin | ||
| valclavam | 0 | 0.7 | 0.49 | 0.79 | 1.0 | Non detected. | MIBiG | Source | CC(C)C(N)C(=O)NC(C(=O)O)C(O)CC1CN2C(=O)CC2O1 | view | valclavam | ||
| Argimycin PV | 1 | 0.65 | 0.48 | 0.69 | 1.0 | Non detected. | MIBiG | Source | C/C=C/C=C1\C=C[C@@H]2NCCC[C@]12O | view | Argimycin PV | ||
| Argimycin PIX | 2 | 0.63 | 0.47 | 0.66 | 1.0 | Non detected. | MIBiG | Source | C/C=C/C=C/CCC1CCCCN1 | view | Argimycin PIX | ||
| Argimycin PVI | 3 | 0.62 | 0.46 | 0.65 | 1.0 | Non detected. | MIBiG | Source | C/C=C/C=C1\C=C[C@@H]2NCCC[C@H]12 | view | Argimycin PVI |